SciML: An Open Source Software Organization for Scientific Machine Learning
Computational scientific discovery is at an interesting juncture. While we have mechanistic models of lots of different scientific phenomena, and reams of data being generated from experiments - our computational capabilities are unable to keep up. Our problems are too large for realistic simulation. Our problems are multiscale and too stiff. Our problems require tedious work like calculating gradients and getting code to run on GPUs and supercomputers. Our next step forward is a combination of science and machine learning, which combines mechanistic models with data based reasoning, presented as a unified set of abstractions and a high performance implementation. We refer to this as scientific machine learning.
Scientific Machine Learning, abbreviated SciML, has been taking the academic world by storm as an interesting blend of traditional scientific mechanistic modeling (differential equations) with machine learning methodologies like deep learning. While traditional deep learning methodologies have had difficulties with scientific issues like stiffness, interpretability, and enforcing physical constraints, this blend with numerical analysis and differential equations has evolved into a field of research with new methods, architectures, and algorithms which overcome these problems while adding the data-driven automatic learning features of modern deep learning. Many successes have already been found, with tools like physics-informed neural networks, deep BSDE solvers for high dimensional PDEs, and neural surrogates showcasing how deep learning can greatly improve scientific modeling practice. At the same time, researchers are quickly finding that our training techniques will need to be modified in order to work on difficult scientific models. For example the original method of reversing an ODE for an adjoint or relying on backpropagation through the solver is not numerically stable for neural ODEs, and traditional optimizers made for machine learning, like Stochastic Gradient Descent and ADAM have difficulties handling the ill-conditioned Hessians of physics-informed neural networks. New software will be required in order to accommodate the unique numerical difficulties that occur in this field, and facilitate the connection between scientific simulators and scientific machine learning training loops.
SciML is an open source software organization for the development and maintenance of a feature-filled and high performance set of tooling for scientific machine learning. This includes the full gamut of tools from differential equation solvers to scientific simulators and tools for automatically discovering scientific models. What I want to do with this post is introduce the organization by explaining a few things:
What SciML provides
What our goals are
Our next steps
How you can join in the process
The Software that SciML Provides
We provide best-in-class tooling for solving differential equations
We will continue to have DifferentialEquations.jl at the core of the organization to support high performance solving of the differential equations that show up in scientific models. This means we plan to continue the research and development in:
Discrete equations (function maps, discrete stochastic (Gillespie/Markov) simulations)
Ordinary differential equations (ODEs)
Split and partitioned ODEs (Symplectic integrators, IMEX Methods)
Stochastic ordinary differential equations (SODEs or SDEs)
Random differential equations (RODEs or RDEs)
Differential algebraic equations (DAEs)
Delay differential equations (DDEs)
Mixed discrete and continuous equations (Hybrid Equations, Jump Diffusions)
(Stochastic) partial differential equations ((S)PDEs) (with both finite difference and finite element methods)
along with continuing to push towards new domains, like stochastic delay differential equations, fractional differential equations, and beyond. However, optimal control, (Bayesian) parameter estimation, and automated model discovery all require every possible bit of performance, and thus we will continue to add functionality that improves the performance for solving both large and small differential equation models. This includes features like:
GPU acceleration through CUDAnative.jl and CuArrays.jl
Automated sparsity detection with SparsityDetection.jl
Automatic Jacobian coloring with SparseDiffTools.jl, allowing for fast solutions to problems with sparse or structured (Tridiagonal, Banded, BlockBanded, etc.) Jacobians
Progress meter integration with the Juno IDE for estimated time to solution
Automatic distributed, multithreaded, and GPU parallelism of ensemble trajectories
Forward and adjoint local sensitivity analysis for fast gradient computations
Built-in interpolations for differential equation solutions
Wrappers for common C/Fortran methods like Sundials and Hairer's radau
Arbitrary precision with BigFloats and Arbfloats
Arbitrary array types, allowing the solution of differential equations on matrices and distributed arrays
We plan to continue our research into these topics and make sure our software is best in class. We plan to keep improving the performance of DifferentialEquations.jl until it is best-in-class in every benchmark we have, and then we plan to add more benchmarks to find more behaviors and handle those as well. Here is a current benchmark showcasing native DifferentialEquations.jl methods outperforming classical Fortran methods like LSODA by 5x on a 20 equation stiff ODE benchmark:
Reference: Pollution Model Benchmarks
We provide tools for deriving and fitting scientific models
It is very rare that someone thinks their model is perfect. Thus a large portion of the focus of our organization is to help scientific modelers derive equations and fit models. This includes tools for:
Forward and adjoint local sensitivity analysis for fast gradients
Some of our newer tooling like DataDrivenDiffEq.jl can even take in timeseries data and generate LaTeX code for the best fitting model (for a recent demonstration, see this fitting of a COVID-19 epidemic model).
We note that while these tools will continue to be tested with differential equation models, many of these tools apply to scientific models in general. For example, while our global sensitivity analysis tools have been documented in the differential equation solver, these methods actually work on any function f(p):
using QuasiMonteCarlo, DiffEqSensitivity
function ishi(X)
A= 7
B= 0.1
sin(X[1]) + A*sin(X[2])^2+ B*X[3]^4 *sin(X[1])
end
n = 600000
lb = -ones(4)*π
ub = ones(4)*π
sampler = SobolSample()
A,B = QuasiMonteCarlo.generate_design_matrices(n,lb,ub,sampler)
res1 = gsa(ishi,Sobol(),A,B)Reorganizing under the SciML umbrella will make it easier for users to discover and apply our global sensitivity analysis methods outside of differential equation contexts, such as with neural networks.
We provide high-level domain-specific modeling tools to make scientific modeling more accessible
Differential equations appear in nearly every scientific domain, but most scientific domains have their own specialized idioms and terminology. A physicist, biologist, chemist, etc. should be able to pick up our tools and make use of high performance scientific machine learning methods without requiring the understanding of every component and using abstractions that make sense to their field. To make this a reality, we provide high-level domain-specific modeling tools as frontends for building and generating models.
DiffEqBiological.jl is a prime example which generates high performance simulations from a description of the chemical reactions. For example, the following solves the Michaelis-Menton model using an ODE and then a Gillespie model:
rs = @reaction_network begin
c1, S + E --> SE
c2, SE --> S + E
c3, SE --> P + E
end c1 c2 c3
p = (0.00166,0.0001,0.1)
tspan = (0., 100.)
u0 = [301., 100., 0., 0.] # S = 301, E = 100, SE = 0, P = 0
# solve ODEs
oprob = ODEProblem(rs, u0, tspan, p)
osol = solve(oprob, Tsit5())
# solve JumpProblem
u0 = [301, 100, 0, 0]
dprob = DiscreteProblem(rs, u0, tspan, p)
jprob = JumpProblem(dprob, Direct(), rs)
jsol = solve(jprob, SSAStepper())This builds a specific form that can then use optimized methods like DirectCR and achieve an order of magnitude better performance than the classic Gillespie SSA methods:
Reference: Diffusion Model Benchmarks
Additionally, we have physics-based tooling and support external libraries like:
NBodySimulator.jl for N-body systems (molecular dynamics, astrophysics)
RigidBodySim.jl for robotics
QuantumOptics.jl for quantum phenomena
DynamicalSystems.jl for chaotic dynamics
We support commercial tooling built on our software like the Pumas software for pharmaceutical modeling and simulation which is being adopted throughout the industry. We make it easy to generate models of multi-scale systems using tools like MultiScaleArrays.jl:
and build compilers like ModelingToolkit.jl that provide automatic analysis and optimization of model code. By adding automated code parallelization and BLT transforms to ModelingToolkit, users of DiffEqBiological, Pumas, ParameterizedFunctions.jl, etc. will all see their code automatically become more efficient.
We provide high-level implementations of the latest algorithms in scientific machine learning
The translational step of bringing new methods of computational science to scientists in application areas is what will allow next-generation exploration to occur. We provide libraries like:
DiffEqFlux.jl for neural and universal differential equations
DataDrivenDiffEq.jl for automated equation generation with Dynamic Mode Decomposition (DMD) and SInDy type methods
ReservoirComputing.jl for echo state networks and prediction of chaotic systems
NeuralPDE.jl for Physics-Informed Neural Networks (PINNs) and Deep BSDE solvers of 100 dimensional PDEs
We will continue to expand this portion of our offering, building tools that automatically solve PDEs from a symbolic description using neural networks, and generate mesh-free discretizers.
We provide users of all common scientific programming languages the ability to use our tooling
While the main source of our tooling is centralized in the Julia programming language, we see Julia as a "language of libraries", like C++ or Fortran, for developing scientific libraries that can be widely used across the whole community. We have previously demonstrated this capability with tools like diffeqpy and diffeqr for using DifferentialEquations.jl from Python and R respectively, and we plan to continue along these lines to allow as much of our tooling as possible be accessible from as many languages as possible. While there will always be some optimizations that can only occur when used from the Julia programming language, DSL builders like ModelingToolkit.jl will be used to further expand the capabilities and performance of our wrappers.
Here's an example which solves stochastic differential equations with high order adaptive methods from Python:
# pip install diffeqpy
from diffeqpy import de
# diffeqpy.install()
def f(du,u,p,t):
x, y, z = u
sigma, rho, beta = p
du[0] = sigma * (y - x)
du[1] = x * (rho - z) - y
du[2] = x * y - beta * z
def g(du,u,p,t):
du[0] = 0.3*u[0]
du[1] = 0.3*u[1]
du[2] = 0.3*u[2]
numba_f = numba.jit(f)
numba_g = numba.jit(g)
u0 = [1.0,0.0,0.0]
tspan = (0., 100.)
p = [10.0,28.0,2.66]
prob = de.SDEProblem(numba_f, numba_g, u0, tspan, p)
sol = de.solve(prob)
# Now let's draw a phase plot
ut = numpy.transpose(sol.u)
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.plot(ut[0,:],ut[1,:],ut[2,:])
plt.show()We provide tools for researching methods in scientific machine learning
Last but not least, we support the research activities of practitioners in scientific machine learning. Tools like DiffEqDevTools.jl and RootedTrees.jl make it easy to create and benchmark new methods and accelerate the publication process for numerical researchers. Our wrappers for external tools like FEniCS.jl and SciPyDiffEq.jl make it easy to perform cross-platform comparisons. Our stack is entirely written within Julia, which means every piece can be tweaked on the fly, making it easy to mix and match Hamiltonian integrators with neural networks to discover new scientific applications. Our issues and chat channel serve as places to not just debug existing software, but also discuss new methods and help create high performance implementations.
In addition, we support many student activities to bring new researchers into the community. Many of the maintainers of our packages, like Yingbo Ma, Vaibhav Dixit, Kanav Gupta, Kirill Zubov, etc. all started as one of our over 50 student developers from past Google Summer of Code and other Julia Seasons of Contributions.
The Goal of SciML
When you read a paper that is mixing neural networks with differential equations (our recent paper, available as a preprint) or designing new neural networks that satisfy incompressibility for modeling Navier-Stokes, you should be able to go online and find tweakable, high quality, and highly maintained package implementations of these methodologies to either start using for your scientific research, or utilize as a starting point for furthering the methods of scientific machine learning. For this reason, the goal of the SciML OSS organization is to be a hub for the development of robust cross-language scientific machine learning software. In order to make this a reality, we as an organization commit to the following principles:
Everything that we build is compatible with automatic differentiation
Putting an arbitrary piece of code from the SciML group into a training loop of some machine learning library like Flux will naturally work. This means we plan to enforce coding styles that are compatible with language-wide differentiable programming tools like Zygote, or provide pre-defined forward/adjoint rules via the derivative rule package ChainRules.jl.
As demonstrated in the following animation, you can take our stochastic differential equation solvers and train a circuit to control the solution by simply piecing together compatible packages.
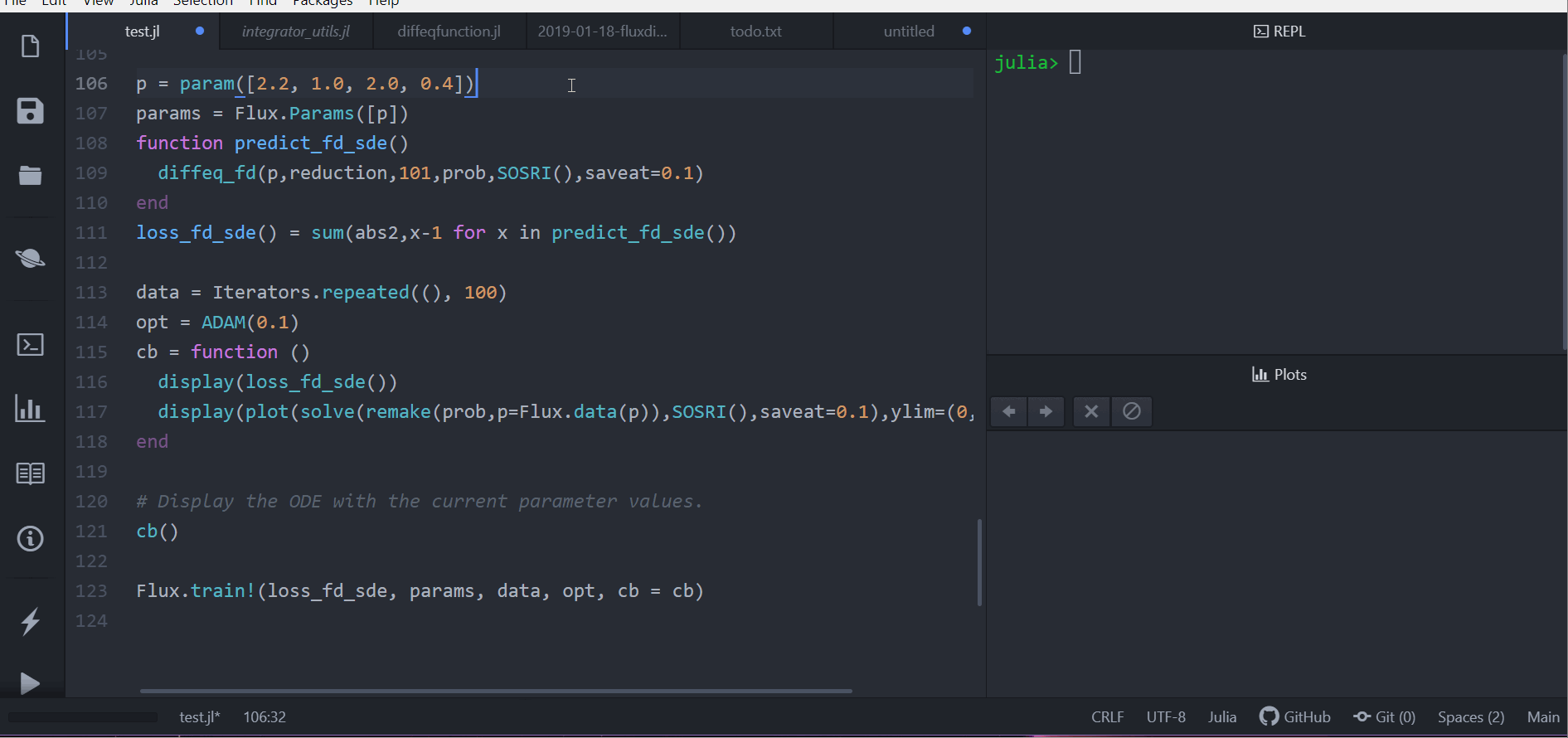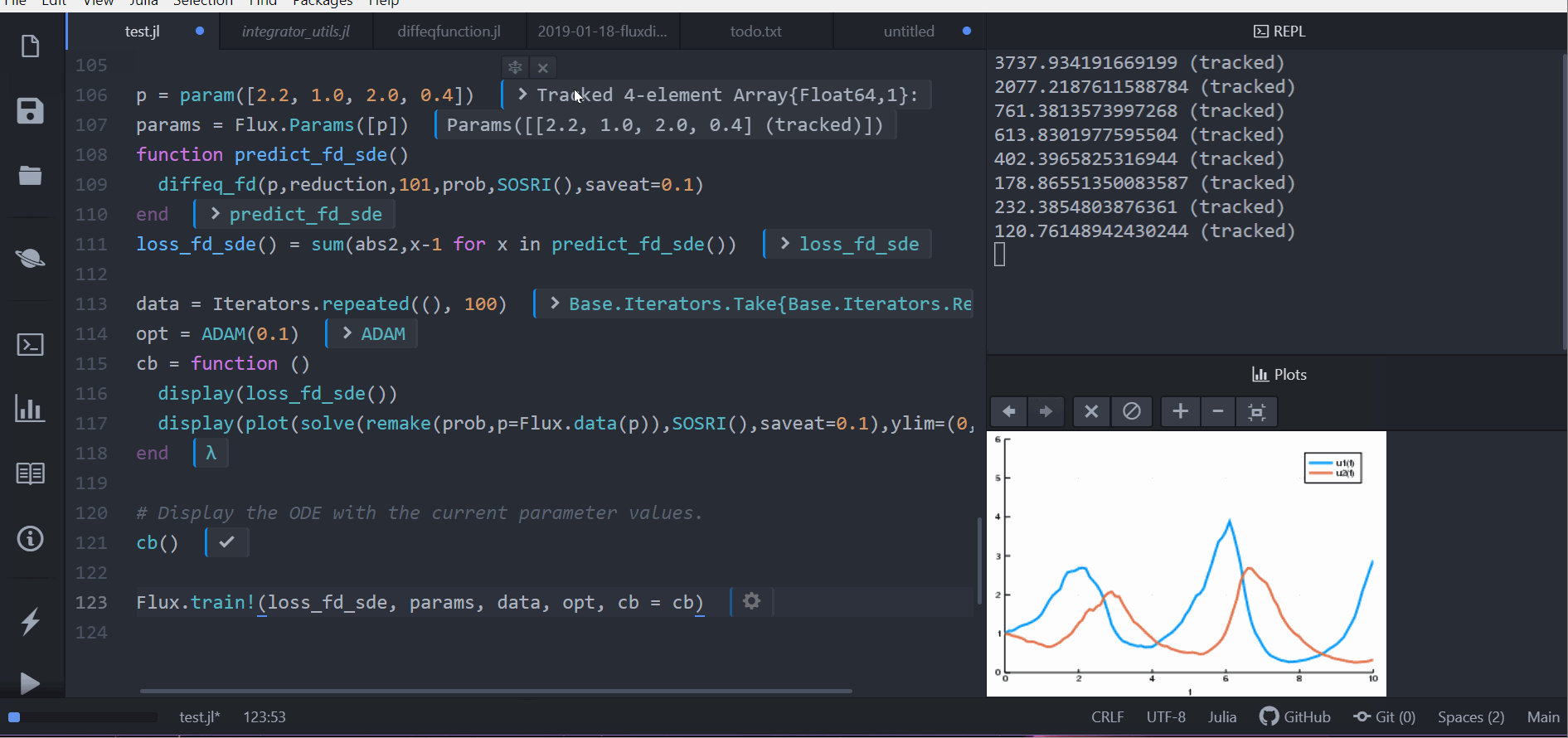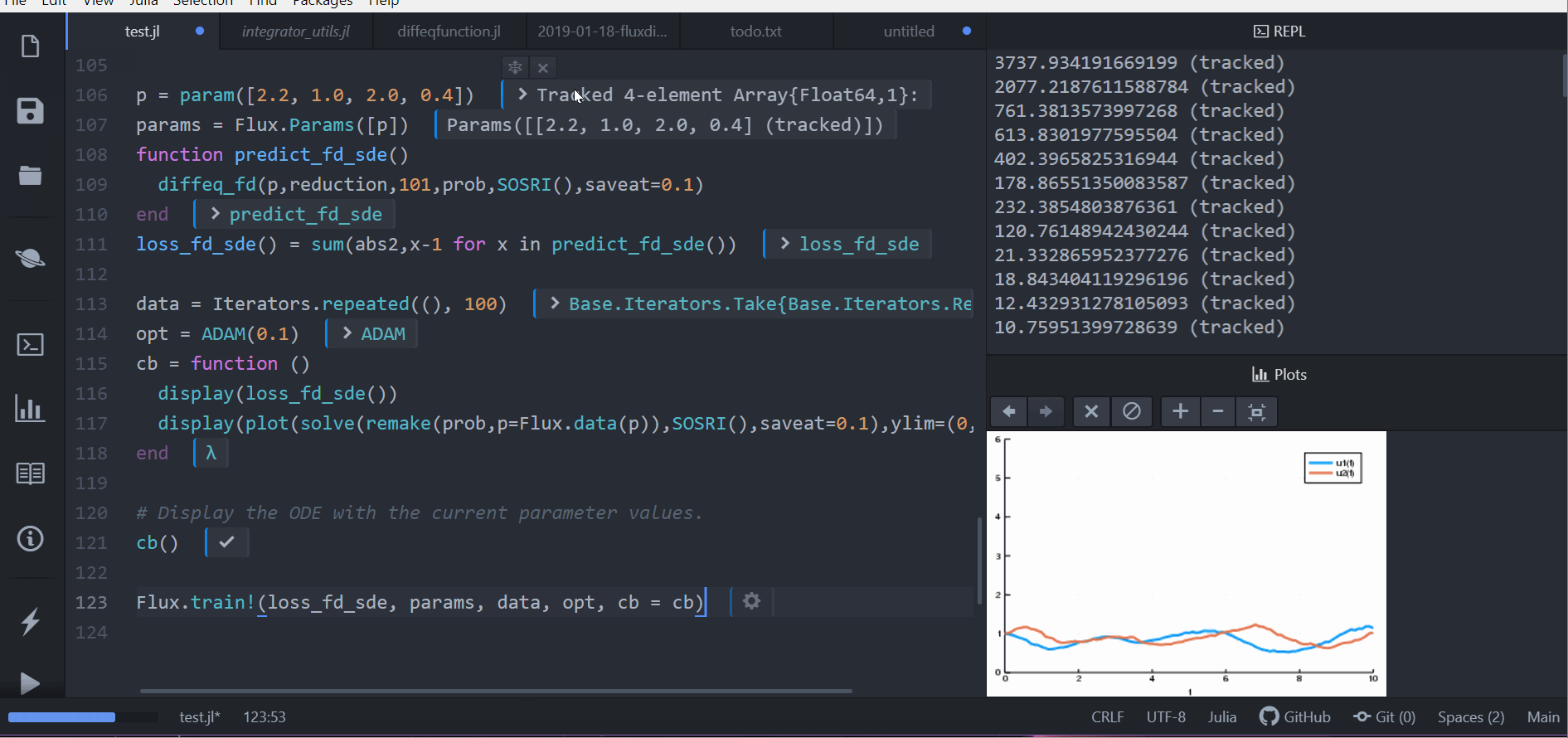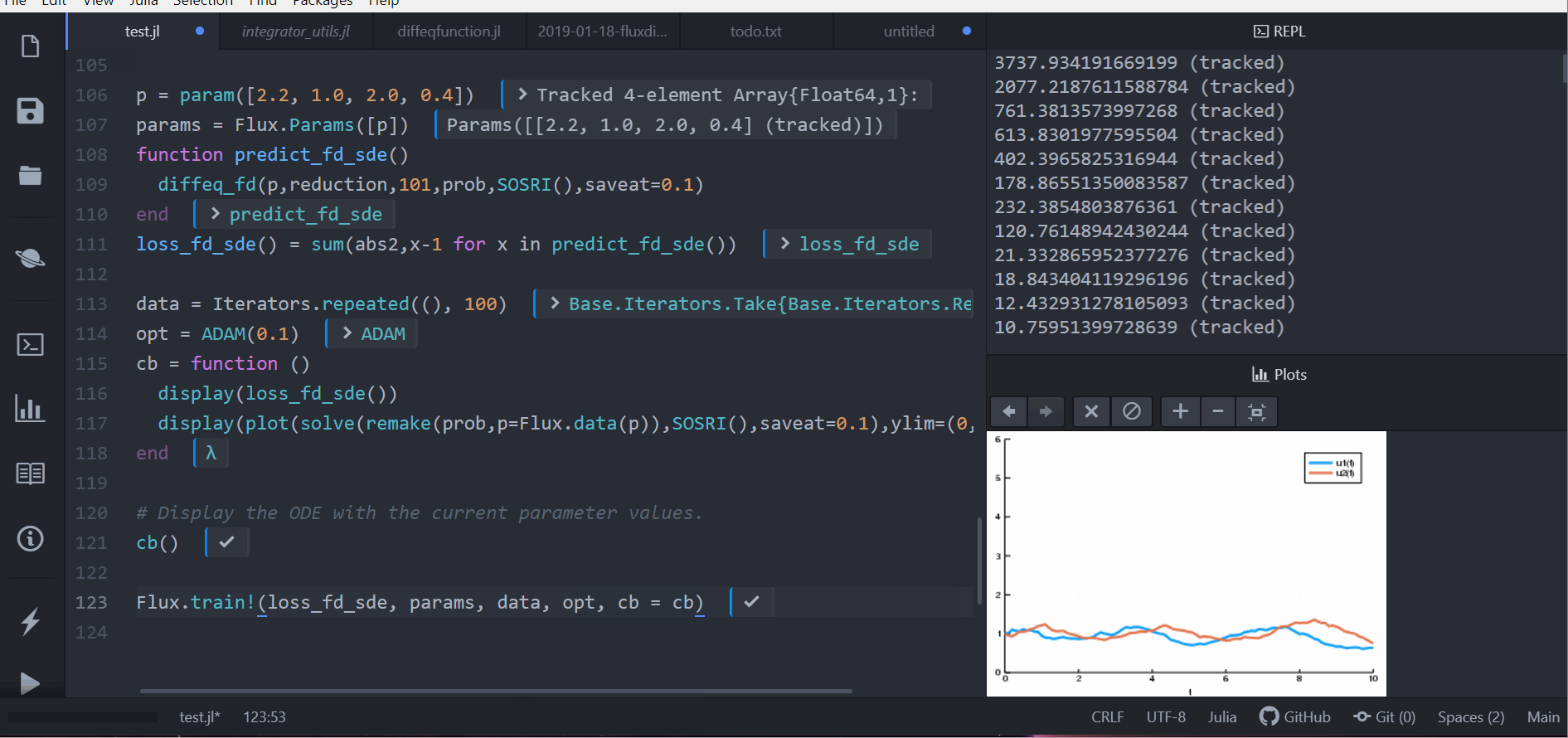
Performance is considered a priority, and performance issues are considered bugs
No questions asked. If you can find something else that is performing better, we consider that an issue and should get it fixed. High performance is required for scientific machine learning to scale, and so we take performance seriously.
Our packages are routinely and robustly tested with the tools for both scientific simulation and machine learning
This means we will continue to develop tools like DiffEqFlux.jl which supports the connection between the DifferentialEquations.jl differential equation solvers and the Flux deep learning library. Another example includes our surrogate modeling library, Surrogates.jl which is routinely tested with DifferentialEquations.jl and the machine learning AD tooling like Zygote.jl, meaning that you can be sure that our surrogates modeling tools can train on differential equations and then be used inside of deep learning stacks. It is this interconnectivity that will allow next-generation SciML methodologies to get productionized in a way that will impact "big science" and industrial use.
We keep up with advances in computational hardware to ensure compatibility with the latest high performance computing tools.
Today, Intel CPUs and NVIDIA GPUs are the dominant platforms, but that won't always be the case. One of the upcoming top supercomputers will be entirely AMD-based, with AMD CPUs and AMD GPUs. In addition, Intel GPUs are scheduled to be a component in future supercomputers. We are committed to maintaining a SciML toolchain that works on all major platforms, updating our compiler backends as new technology is released.
Our Next Steps
To further facilitate our focus to SciML, the next steps that we are looking at are the following:
We will continue to advance differential equation solving in many different directions, such as adding support for stochastic delay differential equations and improving our methods for DAEs.
We plan to create a new documentation setup. Instead of having everything inside of the DifferentialEquations.jl documentation, we plan to split out some of the SciML tools to their own complete documentation. We have already done this for Surrogates.jl. Next on the list is DiffEqFlux.jl which by looking at the README should be clear is in need of its own full docs. Following that we plan to fully document NeuralPDE.jl and its Physics-Informed Neural Networks (PINN) functionality, DataDrivenDiffEq.jl, etc. Because it does not require differential equations, we plan to split out the documentation of Global Sensitivity Analysis to better facilitate its wider usage.
We plan to continue improving the ModelingToolkit ecosystem utilizing its symbolic nature for generic specification of PDEs. This would then be used as a backend with Auto-ML as an automated way to solve any PDE with Physics-Informed Neural Networks.
We plan to continue benchmarking everything, and improve our setup to include automatic updates to the benchmarks for better performance regression tracking. We plan to continue adding to our benchmarks, including benchmarks with MPI and GPUs.
We plan to improve the installation of the Python and R side tooling, making it automatically download precompiled Julia binaries so that way users can utilize the tooling just by using CRAN or pip to install the package. We plan to extend our Python and R offerings to include our neural network infused software like DiffEqFlux and NeuralPDE.
We plan to get feature-completeness in data driven modeling techniques like Radial Basis Function (RBF) surrogates, Dynamic Mode Decomposition and SInDy type methods, and Model Order Reduction.
We plan to stay tightly coupled to the latest techniques in SciML, implementing new physically-constrained neural architectures, optimizers, etc. as they are developed.
How You Can Join in the Process
If you want to be a part of SciML, that's great, you're in! Here are some things you can start doing:
Star our libraries like DifferentialEquations.jl. Such recognition drives our growth to sustain the project.
Join our chatroom to discuss with us.
If you're a student, find a summer project that interests you and apply for funding through Google Summer of Code or other processes (contact us if you are interested)
Start contributing! We recommend opening up an issue to discuss first, and we can help you get started.
Help update our websites, tutorials, benchmarks, and documentation
Help answer questions on Stack Overflow, the Julia Discourse, and other sites!
Hold workshops to train others on our tools.
There are many ways to get involved, so if you'd like some help figuring out how, please get in touch with us.